
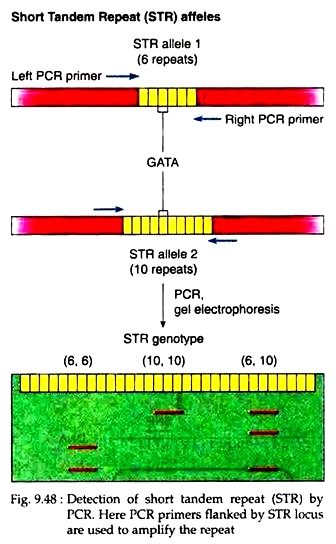
Human genomic variations include single-nucleotide polymorphisms (SNPs), variable number of tandem repeats (VNTRs), short tandem repeats (STRs, or microsatellites), and copy number variations (CNVs). Structural genomic variations also provide important information about both genetic diversity and human evolution. The detection of genomic variations is important in studying the relationships between causative genes and diseases and the relationships between predisposing genes and complex trait diseases, such as type 2 diabetes, coronary heart disease, and cancers.
Our results showed that MSR may be used to (i) physically locate indel and STR sequences and determine STR copy number by searching NCBI reference sequences (ii) predict combinations of microsatellite patterns using the Federal Bureau of Investigation Combined DNA Index System (CODIS) (iii) determine human papilloma virus (HPV) genotypes by searching current viral databases in cases of double infections (iv) estimate the copy number of paralogous genes, such as β-defensin 4 ( DEFB4) and its paralog HSPDP3. The heterozygous sequences are identified as two distinct sequences and aligned with reference sequences. abi file format using comparisons with reference sequences.
BASE SEQUENCE ANALYSIS FREE
In this study, we developed a free web-based program, Mixed Sequence Reader (MSR), which can directly analyze heterozygous base-calling fluorescence chromatogram data in. However, the detection of other genomic variants remains a challenge due to the lack of appropriate tools for heterozygous base-calling fluorescence chromatogram data analysis. Indels and STRs can be easily detected using the currently available In delligent or ShiftDetector programs, which do not search reference sequences. The direct sequencing of PCR products generates heterozygous base-calling fluorescence chromatograms that are useful for identifying single-nucleotide polymorphisms (SNPs), insertion-deletions (indels), short tandem repeats (STRs), and paralogous genes.


 0 kommentar(er)
0 kommentar(er)
